Proposed Foundations for Evaluating Data Sharing and Reuse
in the Biomedical Literature
Biomedical Knowledge Engineering Laboratory
Department of Biomedical Informatics, University of Pittsburgh
200 Meyran Avenue
Pittsburgh, PA 15260
1.412.647.7113
hpiwowar@gmail.com
ABSTRACT
Science progresses by building upon previous research. Progress can be most rapid, efficient, and focused when raw datasets from previous studies are available for reuse. To facilitate this practice, funders and journals have begun to request and require that investigators share their primary datasets with other researchers. Unfortunately, it is difficult to evaluate the effectiveness of these policies. This study aims to develop foundations for evaluating data sharing and reuse decisions in the biomedical literature by developing tools to answer the following research questions, within the context of biomedical gene expression datasets: What is the prevalence of biomedical research data sharing? Biomedical research data reuse? What features are most associated with an investigator’s decision to share or reuse a biomedical research dataset? Does sharing or reusing data contribute to the impact of a research article, independently of other factors? What do the results suggest for developing efficient, effective policies, tools, and initiatives for promoting data sharing and reuse? I suggest a novel approach to identifying publications that share and reuse datasets, through the application of natural language processing techniques to the full text of primary research articles. Using these classifications and extracted covariates, univariate and multivariate analysis will assess which features are most important to data sharing and reuse prevalence, and also estimate the contribution that sharing data and reusing data make to a publication’s research impact. I hope the results will inform the development of effective policies and tools to facilitate this important aspect of scientific research and information exchange.
Categories and Subject Descriptors
H.1.1 [Systems and Information Theory]: Value of information;
H.3.5 [Online Information Services]: Data sharing; J.3 [Life and Medical Sciences]: Biology and genetics, Health
General Terms
Measurement, Human Factors
Keywords
data sharing, data reuse, evaluation, policy, bioinformatics, bibliometrics
1. INTRODUCTION
Sharing information facilitates science. Reusing previously-collected data in new studies allows these valuable resources to contribute far beyond their original analysis.[1] In addition to being used to confirm original results, raw data can be used to explore related or new hypotheses, particularly when combined with other publicly available data sets. Real data is indispensable when investigating and developing study methods, analysis techniques, and software implementations. The larger scientific community also benefits: sharing data encourages multiple perspectives, helps to identify errors, discourages fraud, is useful for training new researchers, and increases efficient use of funding and patient population resources by avoiding duplicate data collection.
Believing that that these benefits outweigh the costs of sharing research data, many initiatives actively encourage investigators to make their data available. Some journals require the submission of detailed biomedical data to publicly available databases as a condition of publication.[2,3] Since 2003, the NIH has required a data sharing plan for all large funding grants and has more recently introduced stronger requirements for genome-wide association studies[4,5]; other funders have similar policies. Several government whitepapers[1,6] and high-profile editorials[7-12] call for responsible data sharing and reuse, large-scale collaborative science is providing the opportunity to share datasets within and outside of the original research projects[13,14], and tools, standards, and databases are developed and maintained to facilitate data sharing and reuse.
Despite these investments of time and money, we do not yet understand the prevalence and patterns of data sharing and reuse, the effectiveness of initiatives, or the costs, benefits, and impact of repurposing biomedical research data.
The goal of this study is to build foundational tools, datasets, and analyses for identifying and evaluating data sharing and reuse decisions within the biomedical literature.
2. FINDINGS AND CHALLENGES IN RESEARCH ON SHARING AND REUSE
This section highlights a few major findings and challenges in research on biomedical data sharing and reuse.
2.1 Understanding Attitudes and Behavior
The largest body of knowledge about motivations and predictors for data sharing and withholding comes from Campbell and co-authors[15-17]. They surveyed researchers, asking whether they have ever requested data and been denied, or themselves denied other researchers from access to data. Results indicated that participation in relationships with industry, mentors’ discouragement of data sharing, negative past experience with data sharing, and male gender were associated with data withholding.[15] In another survey, among geneticists who said they intentionally withheld data related to their published work, 80% said it was too much effort to share the data, 64% said they withheld data to protect the ability of a junior team member to publish, and 53% withheld data to protect their own publishing opportunities.[16]
Occasionally, the administrators of centralized data servers publish feedback surveys of their users. As an example, Ventura[18] reports a survey of researchers who submitted and reviewed microarray studies in the Physiological Genomics journal after its mandatory data submission policy had been in place for two years. Almost all (92%) of authors said that they believed depositing microarray data was of value to the scientific community and about half (55%) were aware of other researchers reusing data from the database.
2.2 Identifying Instances of Data Sharing and Reuse
While surveys have provided insight into sharing and reuse behavior, other issues are best examined by studying the demonstrated behavior of scientists. Unfortunately, observed measurement of data behavior is difficult because of the complexity in identifying all episodes of data sharing and reuse. Although indications of sharing and reuse usually exist within a published research report, the descriptions are in unstructured free text and thus complex to extract.
Data sharing can sometimes be inferred from the “primary citation” field of database submission entries, however these references often missing when data is submitted prior to study publication. Populating the submission citation fields retrospectively requires intensive manual effort, as demonstrated by the recent Protein Data Bank remediation project[19], and thus is not usually performed. No effective way exists to automatically retrieve and index data housed on personal or lab websites or journal supplementary information.
Identifying instances of data reuse is even more difficult. There are few collections or queries that identify studies which reuse data, with the exception of meta-analyses.
Reuse often (but not always) includes a citation reference to the study that produced the data. Mercer et al.[20] is one of several researcher teams who have derived a classification schema for citation contexts. Several groups have methods of automatically classifying citation contexts using natural language processing (NLP) techniques; Teufel et. al[21] uses cue phrases to classify citations into several groups, including a broad “adapts or modifies tools/methods/data” category.
2.3 Estimating the Costs and Benefits of Data Sharing and Reuse
Estimating the costs and benefits of data sharing and reuse would be challenging even with a comprehensive dataset of occurrences. A complete evaluation would require comparing projects that shared or reused with other similar projects that did not, across a wide variety of variables including person-hours-till-completion, total project cost, received citations and their impact, the number and impact of future publications, promotion, success in future grant proposals, and general recognition and respect in the field.
Pienta[22] is currently investigating these questions with respect to social science research data and publications. Zimmerman[23] has studied the ways in which ecologists find and validate datasets to overcome the personal costs and risks of data reuse.
Examining variables for their benefits on research impact is a common theme within the field of bibliometrics. Research impact is usually approximated by citation metrics, despite their recognized limitations.[24]
2.4 Evaluating the Impact of Data Sharing Policies
Studying the impact of data sharing policies is difficult because policies are often confounded with other variables. If, for example, impact factor is positively correlated with a strong journal data sharing policy as well as a large research impact, it is difficult to distinguish the direction of causation. Evaluating data sharing policies would ideally involve a randomized controlled trial, but unfortunately this is impractical.
Despite many funder and journal policies requesting and requiring data sharing, the impact of these policies have only been measured in small and disparate studies. McCain manually categorized the journal “Instruction to Author” statements in 1995.[2] A more recent manual review of gene sequence papers found that, despite requirements, up to 15% of articles did not submit their datasets to Genbank.[25]
2.5 Related Fields
Evaluation of data sharing and reuse behavior is related to a number of other active research fields: code reusability in software engineering, motivation in open source projects and corporate knowledge sharing, tools for collaboration, evaluating research output, the sociological study of altruism, information retrieval, usage metrics, data standards, the semantic web, open access, and open notebook science.
3. RESEARCH QUESTIONS
Within the scope of this project, I plan to address the following questions:
- What is the prevalence of biomedical research data sharing? Biomedical research data reuse?
- What features are most associated with an investigator’s decision to share or reuse a biomedical research dataset?
- Does sharing or reusing data contribute to the impact of a research article, independently of other factors?
- What do the results suggest for developing efficient, effective policies, tools, and initiatives for promoting data sharing and reuse?
I will consider these questions within the context of gene expression microarray data. Microarray data provides a useful environment for investigation: despite being valuable for reuse and costly to collect, is not yet universally shared.
4. PROPOSED METHODOLOGY
I propose to address the research questions by (1) collecting a cohort of articles about gene expression microarray data, (2) developing a system to automatically identify instances of dataset sharing and reuse within the cohort, and (3) analyzing the instances of dataset sharing and reuse for univariate and multivariate predictors. These steps are explained in further detail below.
4.1 Data Collection
The cohort will consist of English-language non-review research articles indexed in PubMed under the MeSH term “gene expression profiling,” published between 2000 and 2007 (21000 articles). Using a combination of automated and manual steps I will obtain the full text of all articles that are available electronically in machine-readable format with a University of Pittsburgh HSLS account. The final article count will depend on the availability of machine-readable articles and permission to download articles in bulk from publisher websites.
For each article, I will record many potentially relevant covariates, including number of authors, sources of funding, MeSH terms related to organism and disease of study, journal impact factor, journal subdiscipline, journal data sharing policy (or lack thereof), and whether the article was originally published by the journal as open-access.
Finally, I will record the citation history of the article from the ISI Web of Science. I will attempt to remove self-citations and reuse citations by investigators who previously co-authored a paper with the original research team. This will hopefully eliminate reuse due to restricted data sharing “behind the scenes” with current and former colleagues and students.
4.2 Data Classification
4.2.1 Criteria for classification
For the purposes of this study, I will consider data “shared” if it is publicly available on the internet. I will use a variety of mechanisms to classify each article as Dataset-Producing or Dataset-NonProducing, Dataset-Sharing or Dataset-NonSharing, and Dataset-Reusing or Dataset-NonReusing.
An article will be considered Dataset-Producing if the full text indicates the execution of a wet-lab gene expression microarray experiment.
All Dataset-Producing articles will be assessed for Dataset-Sharing status. I will consider an article to have shared its dataset if: (a) its PubMed ID or citation is included in a dataset submission record within the Gene Expression Omnibus (GEO)[26], ArrayExpress[27], or SMD[28] databases, or (b) it contains lexical phrases indicating data submission to a database or website.
All cohort articles will be considered as potentially Dataset-Reusing. I will consider an article to have reused a previously shared microarray dataset if: (a) the article’s PubMed ID or citation is included on a list of data reuse studies (such as the partial list of reused GEO-datasets maintained on GEO’s website), (b) the article has MeSH terms suggesting it is a meta-analysis, or (c) the article’s full text contains lexical phrases and/or citations indicating data reuse.
4.2.2 Automatic classification system
I will manually classify a random subset of cohort articles according to the above criteria, and use this gold standard to develop and validate a natural language processing (NLP) systems to do the classifications automatically. The NLP approach will be similar to the preliminary work on Dataset-Sharing classification described in Section 5.4. I also plan on experimenting with additional NLP techniques such as semi-supervised training[29], bootstrapping cue phrases[30], and boosting classifiers as necessary.
I expect the Dataset-Producing classification problem to be relatively straightforward because standard and relatively-specific terms are used to describe the method for a gene expression experiment (e.g., RNA extraction, hybridization, imaging). I expect Dataset-Reuse classification to be fairly challenging, because there are so many diverse ways to acknowledge data reuse provenance within free text.
4.3 Analysis to address Research Questions
4.3.1 Prevalence of sharing and reuse
I will compute the prevalence of sharing by dividing the number of articles identified as Dataset-Sharing by the number identified as Dataset-Producing. The prevalence of reuse is simply the number of articles identified as Dataset-Reusing divided by the total number of articles in the cohort.
4.3.2 Features associated with sharing and reuse
For each of the three cohort classifications (Dataset-Producing, Dataset-Sharing, and Dataset-Reusing), I will compute the univariate odds ratio for each of the covariates described in Section4.1. I will also compute a multivariate logistic regression using these covariates for each of the three classifications.
4.3.3 Effect of sharing and reuse on article impact
I will use regression to assess the association between data decisions and research impact (approximated by citation count), independently of other covariates known to impact citation count. I will compute a multivariate linear regression over the logarithmic-transform of each article’s yearly citation count, including as independent variables the collected covariates, the three binary covariates representing the Dataset-Producing classification, Dataset-Sharing classification, and Dataset-Reusing classification, and interaction terms.
4.3.4 Implications
I will consider the analysis results in light of the current data sharing policy environment to highlight potential implications.
5. PRELIMINARY RESULTS
This project proposal involves integrating and extending the preliminary work described below.
5.1 Data Sharing Impact in a Pilot Cohort
We conducted a preliminary investigation into the citation impact of data sharing by a small, homogeneous cohort of studies.[31] Using linear regression, we found that studies with publicly shared microarray data were associated with a 69% increase in citation count compared to studies without shared datasets, independently of journal impact factor, date of publication, and author country of origin (see Table 1).
Table 1. Multivariate regression on citation count for 85 clinical cancer microarray publications. Reproduced from [31].

The project extends this preliminary analysis by considering a larger and more heterogeneous cohort, additional covariates, an analysis to predict sharing prevalence, and the additional endpoint of dataset reuse.
5.2 Impact of Journal Policy
We conducted a pilot study to understand the current state of data sharing policies within journals, the features of journals that are associated with the strength of their data sharing policies, and whether the strength of data sharing policies impact the observed prevalence of data sharing.[3] We measured data sharing prevalence as the proportion of papers with submission links from NCBI’s Gene Expression Omnibus (GEO) database. We conducted univariate and linear multivariate regressions to understand the relationship between the strength of data sharing policy and journal impact factor, journal subdiscipline, journal publisher (academic societies vs. commercial), and publishing model (open vs. closed access). Of the 70 journal policies, 53 made some mention of sharing publication-related data within their Instruction to Author statements. Of the 40 policies with a data sharing policy applicable to microarrays, we classified 17 as weak and 23 as strong. Policy strength was positively associated with measured data sharing submission into the GEO database: the journals with no data sharing policy, a weak policy, and a strong policy had median data sharing prevalence of 8%, 20%, and 25% respectively (see Figure 1).
This preliminary work suggests that journal policy is an important factor, and (when a policy exists) is extractable from a journal’s Information to Author statement.
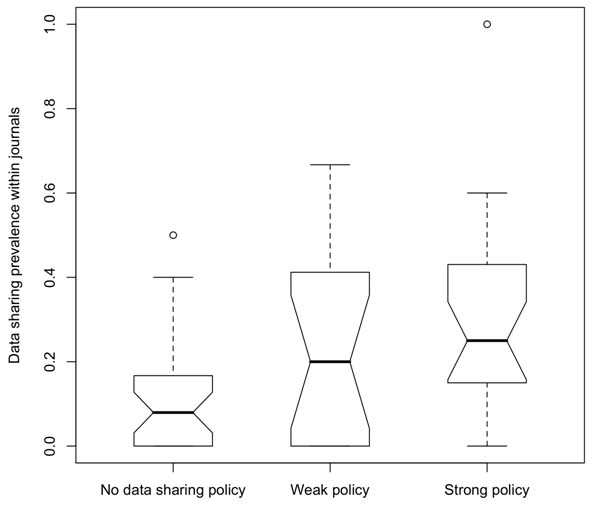
Figure 1. A boxplot of the relative data-sharing prevalence for each journal, grouped by the strength of the journal’s data-sharing policy. For each group, the heavy line indicates the median and the box encompasses the interquartile. NOTE: Prevalence analysis has not been restricted to data-producing articles, and so must be considered relatively and not to an absolute of 100%. Reproduced from [3].
5.3 Prevalence across Research Topic Areas
We performed a preliminary investigation of rough keyword-based Dataset-Producing and Dataset-Reusing classifiers, trained and tested on a manually-labeled set of documents (PLoS articles prior to January 2007 containing the word “microarray,” n=200).[32] We compared the Medical Subject Heading (MeSH) terms of the articles identified as Dataset-Reusing to those identified as Dataset-Producing to estimate the odds that a specific MeSH term would be used given all studies with original microarray data, compared to the odds of the same term describing studies with re-used data. Publications with reused data did involve a relatively high proportion of studies involving fungi (odds ratio (OR)=2.4), and a relatively low proportion involving rats, bacteria, viruses, plants, or genetically-altered or inbred animals (OR<0.5) compared to publications with original data.
We also assessed the prevalence and patterns of Dataset-Sharing, using only links from within the GEO or ArrayExpress database[33]. Of the 2503 articles, 440 (18%) articles had links from either GEO or ArrayExpress. Interestingly, studies with free full text at PubMed were twice (OR=2.1) as likely to be linked as a data source within GEO or ArrayExpress than those without free full text, as seen in Figure 2. Studies with human data were less likely to have a link (OR=0.8) than studies with only non-human data. The proportion of articles with a link within these two databases has increased over time: the odds of a data-source link for studies was 2.5 times greater for studies published in 2006 than 2002. As might be expected, studies with the fewest funding sources had the fewest data-sharing links: only 28 (6%) of the 433 studies with no funding source were listed within GEO or ArrayExpress. In contrast, studies funded by the NIH, the US government, or a non-US government source had data-sharing links in 282 of 1556 cases (18%), while studies funded by two or more of these mechanisms were listed in the databases in 130 out of 514 cases (25%).
These studies demonstrate that funding source, organism, and open-access are important covariates in data behavior.

Figure 2. Preliminary Indications of Data-Sharing Patterns with 95% confidence intervals. Reproduced from [33].
5.4 Automatic Identification of Data Sharing
A pilot NLP system has been developed and validated for one of the three proposed cohort classifications, Dataset-Sharing versus Dataset-NonSharing.[34] Using regular expression patterns and machine learning algorithms on open access biomedical literature published in 2006, our system was able to identify 61% of articles with shared datasets with 80% precision. A simpler version of our classifier achieved higher recall (86%), though lower precision (49%).
These results demonstrate the feasibility of using an NLP approach to automatically identify instances of data sharing from biomedical full text research articles.
6. LIMITATIONS AND RISKS
We note an important limitation of this proposal: associations do not imply causation. The research here will not be sufficient to conclude, for example, that a policy change associated with increased data sharing will in fact cause increased sharing. It would be possible that both factors stem from a common cause.
This study has several other limitations. Although restricting the study to one datatype allows an in-depth analysis of many specific facets of data sharing and reuse, future work should apply the methodology and lessons learned to other datatypes to quantify generalizability. My sample will omit some articles: I might not find all of the datasets that have been shared in niche databases or on personal or lab websites and not all papers will be available in machine-readable full text, particularly for early years. I am not considering datasets as shared if they are available upon request and may thereby discount an important and effective sharing mechanism. Although broadly used, citations are a rough and imperfect measurement of research impact, in part because they may include negative critiques of an article or its associated data. Citations do not consider reuse in the context of education and training, and thus undervalue the impact of data sharing reused for this purpose.
The largest technical risk in the research plan is that it may be unexpectedly difficult to automatically identify reuse with acceptable precision and recall. In this case, I plan to supplement the automated classification with manual curation, possibly resulting in a smaller cohort of articles for analysis.
7. ANTICIPATED CONTRIBUTIONS
I anticipate several important contributions arising from this novel research application.
First, I would, of course, make my dataset publicly available (limited only by licensing restrictions). This would provide a foundational data sharing and reuse dataset for further study by other researchers. I could imagine future work extending and refining my analysis, using the data to investigate novel questions such as whether the data-sharing community has members in common with the data-reuse community –interesting, and also relevant to developing incentives and policies. I envision the reuse data forming the backbone of a Data Reuse Registry, providing a prototype system for ongoing prospective data reuse attribution and cataloguing.[35]
Although some of the analysis results may be intuitive (a stronger journal data sharing policy results in more data sharing, or shared data permits reuse and thus supports a higher citation rate), these relationships have not yet been demonstrated. Concrete, supporting – or contradictory! – evidence will be of value to a wide spectrum of decision-makers.
I hope this research will identify sub-communities with frequent practices of sharing and reuse. Examining these situations can highlight best practices to be used when developing research agendas, tools, standards, repositories, and communities in areas that have yet to receive major benefits from shared data.
Finally, but most importantly, I believe this research will inspire further work in this area. There is a common adage: “You can not manage what you do not measure.” Research consumes considerable resources from the public trust. As data sharing and reuse are evaluated and policies and incentives improved, hopefully investigators will become more apt to share and reuse study data and thus maximize its usefulness to society.
8. ACKNOWLEDGMENTS
I thank my advisor Dr Wendy Chapman for her support and discussion of these ideas, the 2008 Joint Conference on Digital Libraries Doctoral Consortium reviewers and participants for their insightful feedback, Virginia Tech for travel funding, and the NLM for training support through grant 5T15-LM007059-19.
9. REFERENCES
| [1] | S. E. Fienberg, M. E. Martin, and M. L. Straf, Sharing Research Data, Washington, D.C.: National Academy Press, 1985. |
| [2] | K. McCain, "Mandating Sharing: Journal Policies in the Natural Sciences," Science Communication, vol. 16, no. 4, pp. 403-431, 1995. |
| [3] | H. A. Piwowar and W. W. Chapman, "A Review of Journal Policies for Sharing Research Data," in ELPUB 2008. Available: http://elpub.scix.net/cgi-bin/works/Show?001_elpub2008 |
| [4] | "NIH Data Sharing Policy and Implementation Guidance." Available: http://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm |
| [5] | "NOT-OD-08-013: Implementation Guidance and Instructions for Applicants: Policy for Sharing of Data Obtained in NIH-Supported or Conducted Genome-Wide Association Studies (GWAS), " NIH. Available: http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-013.html |
| [6] | T. Cech, Sharing Publication-Related Data and Materials: Responsibilities of Authorship in the Life Sciences, Washington, D.C.: National Academies Press, 2003. |
| [7] | "Got Data?" Nat Neurosci, vol.10, no.8, p. 931, Aug. 2007. |
| [8] | "Compete, Collaborate, Compel", Nat Genet., vol. 39, no. 8, pp.931, Aug. 2007. |
| [9] | "Democratizing Proteomics Data," Nature Biotechnology, vol. 25, no. 3, pp. 262, 2007. |
| [10] | "Time for Leadership," Nature Biotechnology, vol. 25, no. 8, p. 821, 2007. |
| [11] | "How to Encourage the Right Behaviour," Nature, vol. 416, no. 6876, p.1, 2002. |
| [12] | R.B. Altman, et al., "Genetic Nondiscrimination Legislation: a Critical Prerequisite for Pharmacogenomics Data Sharing," Pharmacogenomics, vol. 8, no. 5, p. 519, May 2007. |
| [13] | K. K. Kakazu, L. W. Cheung, and W. Lynne, "The Cancer Biomedical Informatics Grid (caBIG): Pioneering an Expansive Network of Information and Tools for Collaborative Cancer Research," Hawaii Med J., vol. 63, no. 9, pp. 273-75, Sep. 2004. |
| [14] | GAIN Collaborative Research Group, "New Models of Collaboration in Genome-wide Association Studies: the Genetic Association Information Network," Nat Genet., vol. 39, no .9, pp. 1045-1051, 2007. |
| [15] | D. Blumenthal, E. G. Campbell, M. Gokhale, R. Yucel, B. Clarridge, S. Hilgartner, and N. A. Holtzman, "Data Withholding in Genetics and the Other Life Sciences: Prevalences and Predictors," Acad Med., vol. 81, no. 2, pp.137-145, 2006. |
| [16] | E.G. Campbell, B.R.Clarridge, M. Gokhale, L. Birenbaum, S. Hilgartner, N.A. Holtzman, and D. Blumenthal, "Data Withholding in Academic Genetics: Evidence from a National Survey," JAMA, vol. 287, no. 4, pp. 473-480, 2002. |
| [17] | C. Vogeli, R. Yucel, E. Bendavid, L. M. Jones, M. S.Anderson, K. S. Louis, and E. G. Campbell, "Data Withholding and the Next Generation of Scientists: Results of a National Survey," Acad Med., vol. 81, no. 2, pp.128-36, 2006. |
| [18] | B. Ventura, "Mandatory Submission of Microarray Data to Public Repositories: How is it Working?" Physiol Genomics, vol. 20, no. 2, pp.153-156, 2005. |
| [19] | PDBj, "Report on the Remediation of Primary Citations of PDB data," PDBj News Letter, vol.7, Mar. 2006. Available:
|
| [20] | R. Mercer and C. Di Marco, "The Importance of Fine-Grained Cue Phrases in Scientific Citations," in AI’2003, Proceedings of the 16th Conference of the Canadian Society for the Computational Studies of Intelligence (CSCSI), Halifax, Canada, June 2003. Available: http://ai.uwaterloo.ca/cdimarco/pdf/publications/CSCSI2003.pdf |
| [21] | S. Teufel, A. Siddharthan, and D. Tidhar, "Automatic Classification of Citation Function," in Proc. of EMNLP (ACL 2006), pp.103-110, 2006. |
| [22] | A. Pienta, 1R01LM009765-01 Barriers and Opportunities for Sharing Research Data. NIH Grant. 2007. |
| [23] | A. Zimmerman, "Not by Metadata Alone: the Use of Diverse Forms of Knowledge to Locate Data for Reuse," International Journal on Digital Libraries, vol.7, no.1-2, pp. 5-16, 2007. |
| [24] | P. O. Seglen, "Why the Impact Factor of Journals Should not be Used for Evaluating Research," BMJ., vol. 314, no. 7079, pp. 498-502, 1997. |
| [25] | M. A. Noor, K. J. Zimmerman, and K. C. Teeter, "Data Sharing: How Much Doesn't Get Submitted to GenBank?" PLoS Biol., vol. 4, no.7, 2006. |
| [26] | R. Edgar, M. Domrachev, and A. E. Lash, "Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository," Nucleic Acids Res., vol. 30, no. 1, pp. 207-210, Jan. 2002. |
| [27] | A. Brazma, H. Parkinson, U. Sarkans, M. Shojatalab, J. Vilo, N. Abeygunawardena, E. Holloway, M. Kapushesky, P. Kemmeren, G. G. Lara, A. Oezcimen, P. R. Serra, and S. A. Sansone, "ArrayExpress--a public repository for microarray gene expression data at the EBI," Nucleic Acids Research, vol.33, Database issue, pp. D553-555, Jan. 2005. |
| [28] | G. Sherlock, T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J.C. Matese, S.S. Dwight, M. Kaloper, S. Weng, H. Jin; C.A. Ball, M.B. Eisen, P.T. Spellman, P.O. Brown, D. Botstein, and J.M. Cherry, "The Stanford Microarray Database," Nucleic Acids Research, vol.29, no.1, pp.152-155, Jan. 2001. |
| [29] | B. Medlock, "Exploring Hedge Identification in Biomedical Literature," Journal of Biomedical Informatics, vol. 41, no. 4, pp. 636-654, 2008. |
| [30] | R.M. Abdalla and S. Teufel, "A Bootstrapping Approach to Unsupervised Detection of Cue Phrase Variants," in Annual Meeting of the ACL, 2006. |
| [31] | H. A. Piwowar, R. S. Day, and D. B. Fridsma, "Sharing Detailed Research Data is Associated with Increased Citation Rate," PLoS ONE., vol.2, no.3, e308, 2007. |
| [32] | H. A. Piwowar and D. B. Fridsma, "Examining the Uses of Shared Data," [Poster at ISMB 2007]. Available from Nature Precedings: http://dx.doi.org/10.1038/npre.2007.425.3 |
| [33] | H. A. Piwowar and W. W. Chapman, "Prevalence and Patterns of Microarray Data Sharing," [Poster at PSB 2008], March 2008. Available from Nature Precedings: http://dx.doi.org/10.1038/npre.2008.1701.1 |
| [34] | H. A. Piwowar and W. W. Chapman, "Identifying Data Sharing in Biomedical Literature," August 2008. Available from Nature Precedings: http://precedings.nature.com/documents/1721/version/2 |
| [35] | H. A. Piwowar and W. W.Chapman, "Envisioning a Biomedical Data Reuse Registry," [Poster at AMIA 2008]. Available: http://researchremix.wordpress.com/2008/03/24/envisioning-a-biomedical-data-reuse-registry/ |

